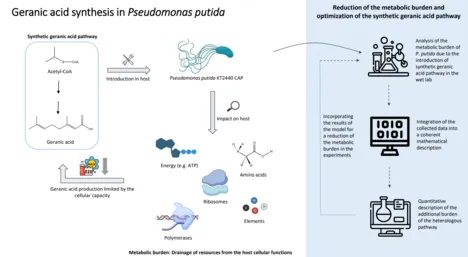
Metabolic burden and resource allocation in Pseudomonas putida during the expression of a synthetic pathway for geranic acid production
Fossil raw materials are still primarily used to produce industrially relevant chemicals. Biotechnological production of these chemicals by bacteria offers a sustainable alternative. However, implementing a heterologous metabolic pathway in a bacterial host increases the demand for energy and cellular resources such as amino acids, ribosomes, and polymerases. The result is a redistribution of normal cellular functions to the additional implemented task.
Restricting even one of these resources usually leads to a reduction in growth rate and cessation of heterologous production, a condition defined as metabolic burden.
A comprehensive understanding of the physiology and metabolism of host heterologous protein overproduction is essential to minimize metabolic stress. This understanding is lacking for Pseudomonas putida, a promising model organism for modern biotechnological applications.
Therefore, this project aims to investigate the metabolic burden of P. putida during heterologous production of geranic acid. Geranic acid is a monoterpenoid with great potential for various industrially relevant applications, e.g., as a fragrance or antifungal agent.
The first step is introducing a synthetic geranic acid metabolic pathway into P. putida. For this, genes from Myxococcus xanthus and Ocimum basilicum are implemented. To follow the cellular capacity during the expression of heterologous proteins, P. putida CAP is applied, which carries the gene for the fluorescent mCherry protein under the control of a constitutive promoter in the chromosome. The mCherry signal indicates cellular capacity, as it reflects the fraction of cellular resources used for non-regulated gene expression. The capacity monitor and the growth rate contribute to the characterization of the metabolic burden. In addition, metabolomic, transcriptomic, and proteomic determinations will be performed to obtain a complete quantitative physiological picture of the manipulated strains.
The collected data will then be integrated into a coherent mathematical description. To understand the cellular processes and constraints during heterologous protein production, it is necessary to quantitatively describe the distribution of resources among the intrinsic functions required for cell maintenance and the additional load imposed by the heterologous pathway. A quantitative description is usually based on a mathematical model, which will be developed in this project. The model can be refined step by step through further experiments and thus passes through the typical life cycle of model development (model improvement - model validation - model application).
Project supervisor: M.Sc. Carina Meiners
Start of the project: 01.11.2022